Access this dataset through Globus. Follow the instructions here.
Data Dictionary v1
Table of contents
Dataset Changes
v1.0.1 Changes (Mar 2024)
- The 12 “short” leads were added to the dataset with a sample rate fo 500 Hz.
- The 3 “long” leads with a sample rate of 500 Hz were added.
File Tree
.
└── ed-bwh-ecg
└── v1
├── ecg-ed-enc.csv
├── ecg-metadata.csv
├── ecg-npy-index.csv
├── ed-encounter.csv
├── ecg-waveform.h5
├── ecg-waveform.npy
└── patient.csv
Entity Relationship Diagram
erDiagram
PATIENT {
string patient_ngsci_id PK
string demographics
}
ECG-METADATA {
string ecg_id PK
string patient_ngsci_id FK
string data
}
ECG-NPY-INDEX {
string ecg_id PK,FK
int npy_index
}
ED-ENCOUNTER {
string enc_id PK
string patient_ngsci_id FK
string data
}
ECG-ED-ENC {
string ecg_id PK,FK
string enc_id FK
}
ECG-WAVEFORM {}
PATIENT ||--|{ ECG-METADATA : patient_ngsci_id
PATIENT ||--|{ ED-ENCOUNTER : patient_ngsci_id
ECG-METADATA ||--|| ECG-NPY-INDEX : ecg_id
ECG-METADATA ||--o| ECG-ED-ENC : ecg_id
ED-ENCOUNTER ||--|{ ECG-ED-ENC : enc_id
ECG-NPY-INDEX ||--|| ECG-WAVEFORM : npy_index
Patient
patient.csv contains patient
| Column Name | Description | Data Type | Example |
|---|---|---|---|
| patient_ngsci_id | Unique patient identifier Pattern: pat{8 digit hex}
| string | pat089c033f |
| sex | Patient Sex | string | Female |
| black | Patient Race/Ethnicity - The patient will only have 1 in one of the categories.
| int | 1 |
| hispanic | int | 0 | |
| white | int | 0 | |
| other | int | 0 | |
| agi_under_25k | Adjusted Gross Income (AGI) distributions from block-level census data based on patient address
| float | 0.36921 |
| agi_25k_to_50k | float | 0.28902 | |
| agi_50k_to_75k | float | 0.15070 | |
| agi_75k_to_100k | float | 0.07804 | |
| agi_100k_to_200k | float | 0.09849 | |
| agi_above_200k | float | 0.01453 |
- Rows: 44,713
- Columns: 12
ECG Waveforms
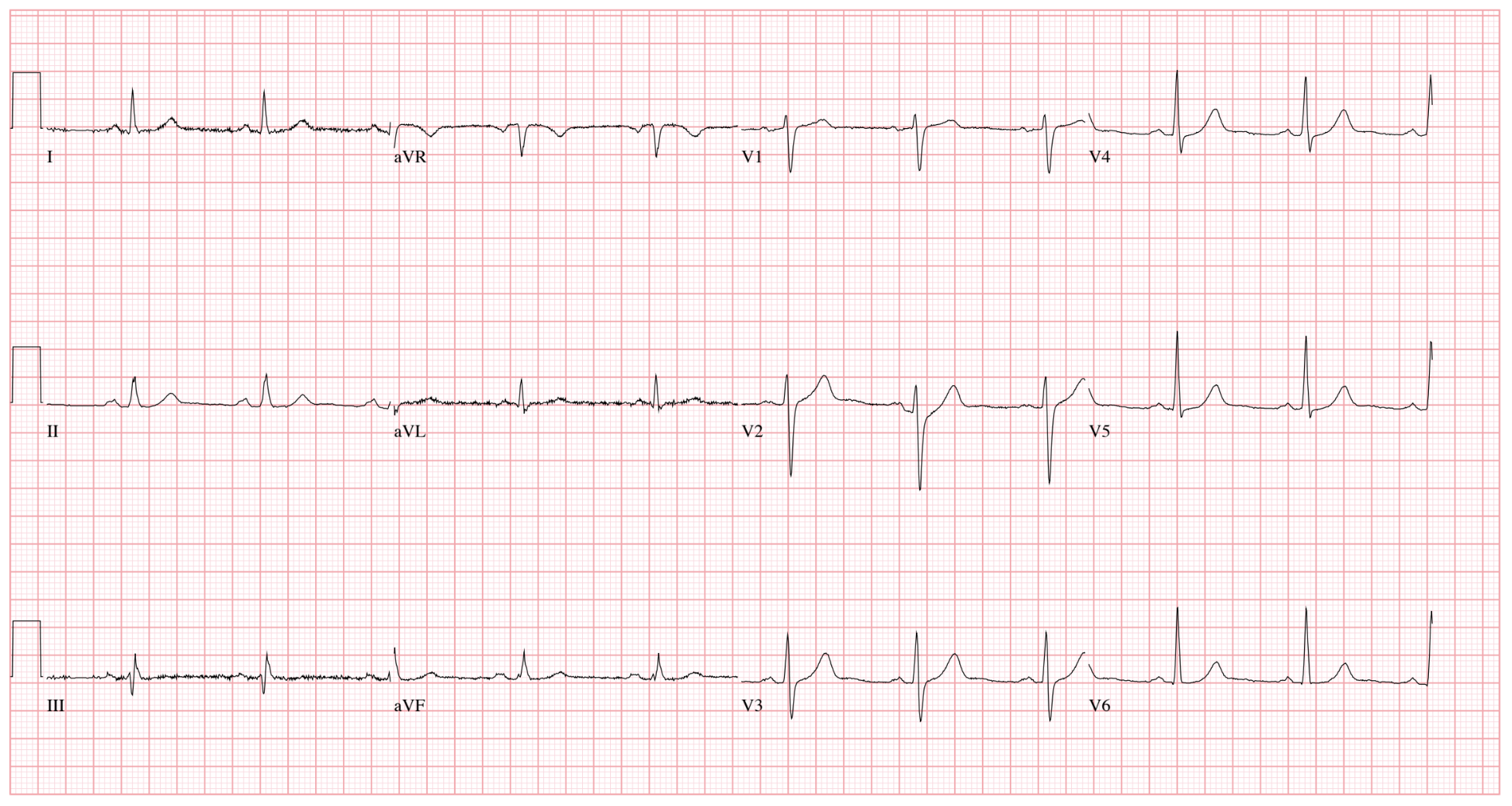
The ECGs for this dataset were collected as PDF files. The waveforms were visibly rendered on the PDF files and needed to be processed in order to be converted into numeric form. The images appeared as in Figure 2.
Long Leads - 100 Hz
The waveforms for leads V1, II, and V5 were smoothed with a rolling average and the sample rate was reduced to 100 Hz. The time duration for these leads is 10 seconds.
ecg-npy-index.csv
| Column Name | Description | Data Type | Example |
|---|---|---|---|
| ecg_id | Unique ECG identifier Pattern: ecg{10 digit hex}
| string | ecg3df45120a4 |
| npy_index | Index of the NumPy array | int | 523 |
- Rows: 112,900
- Columns: 2
ecg-waveform.npy and ecg-waveform.h5
Shape: (112900, 3, 1000)
All Leads - 500 Hz
ed-bwh-ecg/v1/ecg-waveforms-npz/{first two digits of ecg_id}/{ecd_id}.npz
We have also included the numeric waveforms for all leads. These waveforms have a sample rate of 500 Hz.
The waveforms are stored as compressed NumPy files. There is a separate file for each ECG, with the ECG ID in the file name. The array contained in each file has the dimensions of 15 x 5000. The first 12 rows are the waveforms for the 12 leads. However, each of these leads only have a duration of about 2.5 seconds. As seen in Figure 2, leads I, II, and III are measured for the first 2.5 seconds. Leads aVR, aVL, and aVF are measured for the second 2.5 seconds. Leads V1, V2, and V3 are measured for the third 2.5 seconds. And leads V4, V5, and V6 are measured for the last 2.5 seconds. Not seen in Figure 2, leads V1, II, and V5 are measured for the full 10 seconds. Those represent the last 3 rows of the array.
A notebook interacting with these files can be found in the supplementary directory. ed-bwh-ecg/supplementary/notebooks/accessing-waveforms.ipynb
ECG Metadata
Date Shift
Dates in this dataset have been shifted by a random amount for each patient. This is done to create anonymity while preserving the temporal relationship between events for patients.
ecg-metadata.csv
| Column Name | Description | Data Type | Example |
|---|---|---|---|
| patient_ngsci_id | Unique patient identifier Pattern: pat{8 digit hex}
| string | pat089c033f |
| ecg_id | Unique ECG identifier Pattern: ecg{10 digit hex}
| string | ecg3df45120a4 |
| date | Shifted date and time of the ECG | string | 2110-07-29T11:27:56Z |
| p-r-t_axes | P-R-T axes | string | 52 9 27 |
| p_axes | P axes | int | 52 |
| r_axes | R axes | int | 9 |
| t_axes | T axes | int | 27 |
| pr_interval | PR interval | int | 176 |
| pr_interval_units | PR interval units | string | ms |
| qrs_duration | QRS duration | int | 74 |
| qrs_duration_units | QRS duration units | string | ms |
| qtqtc | QTQTc | string | 432/413 ms |
| qt_interval | QT interval | int | 432 |
| qt_interval_units | QT interval units | string | ms |
| qtc_interval | QTc interval | int | 413 |
| qtc_interval_units | QTc interval units | string | ms |
| vent_rate | Vent rate | int | 55 |
| vent_rate_units | Vent rate units | string | BPM |
| has_bbb | Flags for whether search terms were present in the cardiology remarks
| int | 0 |
| has_afib | int | 0 | |
| has_st | int | 0 | |
| has_pacemaker | int | 0 | |
| has_lvh | int | 0 | |
| has_normal | int | 1 | |
| has_normal_ecg | int | 1 | |
| has_normal_sinus | int | 0 | |
| has_depress | int | 0 | |
| has_st_eleva | int | 0 | |
| has_twave | int | 0 | |
| has_aberran_bbb | int | 0 | |
| has_jpoint_repol | int | 0 | |
| has_jpoint_eleva | int | 0 | |
| has_twave_inver | int | 0 | |
| has_twave_abnormal | int | 0 | |
| has_nonspecific | int | 0 | |
| has_rhythm_disturbance | int | 0 | |
| has_prolonged_qt | int | 0 | |
| has_lead_reversal | int | 0 | |
| has_poor_or_quality | int | 0 |
- Rows: 112,900
- Columns: 39
Columns with names that start with has_ indicate whether certain search terms were present in the cardiology remarks. Below are the search terms each flag label.
| Column Name | Regex Search Terms |
|---|---|
| has_bbb |
bbb or bundle\s+branch\s+block
|
| has_afib |
atrial\s+flutter or atrial\s+fibrillation
|
| has_st | st\s+ |
| has_pacemaker |
pacemaker or paced
|
| has_lvh |
lvh or ventricular\s+hypertrophy
|
| has_normal | (normal\s+sinus\s+rhythm and not abnormal\s+sinus\s+rhythm)or ( normal\s+ecg and not abnormal+ecg) |
| has_normal_ecg |
normal\s+ecg and not abnormal\s+ecg
|
| has_normal_sinus |
normal\s+sinus\s+rhythm and not abnormal\s+sinus\s+rhythm
|
| has_depress | st\s*\w*\s*depress |
| has_st_eleva | st\s*\w*\s*eleva |
| has_twave | t.wave |
| has_aberran_bbb |
bbb or bundle\s+branch\s+block or aberran
|
| has_jpoint_repol |
j\s+point or early repol
|
| has_jpoint_eleva |
st\s*\w*\s*eleva or j\s+point or early repol
|
| has_twave_inver |
t.wave and inter
|
| has_twave_abnormal | t.wave.abnormal |
| has_nonspecific | nonspecific |
| has_rhythm_disturbance |
premature (atrial|ventricular)|PAC|PVC or aberran or intraventricular conduction or ectop or arrythmia or junctional or fusion complex or a-v|atrioventricular
|
| has_prolonged_qt | prolonged qt |
| has_lead_reversal | lead reversal |
| has_poor_or_quality |
poor or quality
|
Emergency Department Encounters
Date Shift
Dates in this dataset have been shifted by a random amount for each patient. This is done to create anonymity while preserving the temporal relationship between events for patients.
ed-encounter.csv
| Column Name | Description | Data Type | Example |
|---|---|---|---|
| patient_ngsci_id | Unique patient identifier Pattern: pat{8 digit hex}
| string | pat089c033f |
| ed_enc_id | Unique ED encounter identifier Pattern: enc{8 digit hex}
| string | enc5ba023af |
| start_datetime | Shifted start of the date and time of the ED encounter | string | 2110-07-29T11:06:00Z |
| end_datetime | Shifted end of the date and time of the ED encounter | string | 2110-07-29T12:31:00Z |
| age_at_admit | Patient age | int | 75 |
| macetrop_030_pos | Major adverse cardiovascular events (MACE) & pos troponin in 30 days after visit | bool | FALSE |
| death_030_day | Death in 30 days after visit - This variable comes from Social Security Death Index data, so it captures both death in and out of the hospital. | bool | FALSE |
| macetrop_pos_or_death_030 | Adverse Events (30days) | bool | FALSE |
| stent_010_day | Stent within 10 days after visit | bool | FALSE |
| cabg_010_day | Coronary artery bypass graft surgery (CABG) within 10 days after visit | bool | FALSE |
| stent_or_cabg_010_day | Stent or CABG within 10 days | bool | FALSE |
| ami_day_of | Acute myocardial infarction “heart attack” day of visit days_to_ami == 0
| bool | FALSE |
| days_to_ami | Number or days to soonest AMI, missing if no AMI | int | 5 |
| maxtrop_sameday | Max troponin lab results on day of visit | float | 0.25 |
| tn_group_sameday | Categorized maxtrop_sameday into following bins - missing- 0- (0-0.05]- (0.05-0.1]- (0.1-05]- >0.5
| string | (0.1,0.5] |
| disch_disp | Discharge code | string | a |
| disch_obs | Flag for whether patient is dispatched to observation (disch_disp == e | disch_disp == edobs)
| bool | FALSE |
| test_010_day | Stress test or cath test within 10 days after visit | bool | FALSE |
| stress_010_day | Stress testing (10days) | bool | FALSE |
| cath_010_day | Catheterization (10days) | bool | FALSE |
| days_to_stress | Days to earliest stress test | int | 1 |
| days_to_cath | Days to earliest catheterization test | int | 2 |
| first_test | Whether earliest test is stress or cath; if they have both we generally assume first test is stress even if this doesn’t seem true by timestamps | string | cath |
| excl_flag_c_int | Flag for cardiac intervention in previous 30d | bool | FALSE |
| excl_flag_chronic | Flag for chronic illness | bool | FALSE |
| excl_flag_death | Flag for discharge = death | bool | FALSE |
| exclude_modeling | Exclusion flag for training models = (excl_flag_c_int | excl_flag_chronic | excl_flag_death | (ami_day_of & !test_010_day))
| bool | FALSE |
| exclude | Exclusion flag for analysis = (exclude_modeling | age_at_admit >= 80 | (!test_010_day & maxtrop_sameday > 0))
| bool | FALSE |
- Rows: 71,460
- Columns: 29
ecg-ed-enc.csv
| Column Name | Description | Data Type | Example |
|---|---|---|---|
| ecg_id | Unique ECG identifier Pattern: ecg{10 digit hex}
| string | ecg3df45120a4 |
| ed_enc_id | Unique ED encounter identifier Pattern: enc{8 digit hex}
| string | enc5ba023af |
- Rows: 103,952
- Columns: 2